Abstract
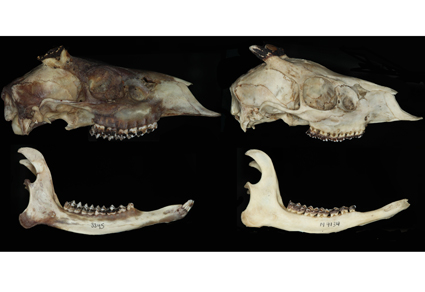
The dwarf red brocket, Mazama rufina (Pucheran, 1851) is a small deer with a fragmented distribution in the montane forests of the Andes of Peru, Ecuador, Colombia, and Venezuela. Little is known about the phylogenetic relationships and the haplotype diversity of its populations, which show distribution gaps. Here we elucidate the phylogenetic relationships of M. rufina and other neotropical deer using mitochondrial data, and analyze genetic geographic variation of this taxon by using haplotype networks of the Cyt-b gene from northern South America. Our analyses recovered M. rufina as independent clade that is not part of Mazama, and sister to a clade composed of Mazama species (except Mazama chunyi) and Odocoileus. The morphometric data of cranial traits confirms that the dwarf red brocket is among the smallest species of deer in South America, only overlapping with small cis-Andean gray brockets (genus Passalites). Based on these results, we provide a new generic classification for this taxon by placing the dwarf red brocket in a new genus found only in the Andes of northern South America. The Cyt-b haplotype network of the dwarf red brocket showed a strong geographic structure caused by the interplay of Cordilleras and lowland river valleys. The genetic distances between the geographic groups were between 1.4 % (Central Cordillera of Colombia vs. Andes of Ecuador) to 2.52 % (Mérida Cordillera vs. Ecuador). The species range using Extent of Occurrence (EOO) and Area of Occupancy (AOO) was 443,764 and 796 km2, respectively, suggesting that the species could be listed as Near Threatened. However, additional information on population changes and susceptibility to habitat transformation is crucial to evaluate whether the dwarf red brocket can be deemed Vulnerable along its distribution. Compared with previous distribution hypotheses, the revised map suggests less extensive distribution gaps in Colombia and highlights priority areas for future sampling in Colombia, Ecuador, and Venezuela.
References
- Aiello‐Lammens, M.E., Boria, R.A., Radosavljevic, A., Vilela, B. & Anderson, R.P. (2015) spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography, 38 (5), 541–545. https://doi.org/10.1111/ecog.01132
- Allen, J.A. (1915) Notes on American deer of the genus Mazama. Bulletin American Museum of Natural History, 34, 521–553.
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
- Bachman, S., Moat, J., Hill, A.W., de la Torre, J. & Scott, B. (2011) Supporting Red List threat assessments with Geo CAT: geospatial conservation assessment tool. ZooKeys, 150, 117–126. https://doi.org/10.3897/zookeys.150.2109
- Barrio, J., Gutiérrez, E.E. & D’Elía, G. (2024) The first living cervid species described in the 21st century and revalidation of Pudella (Artiodactyla). Journal of Mammalogy, 105, 577–588. https://doi.org/10.1093/jmammal/gyae012
- Bernegossi, A.M., Borges, C.H. de S., Sandoval, E.D.P., Cartes, J.L., Cernohorska, H., Kubickova, S., Vozdova, M., Caparroz, R., González, S. & Duarte, J.M.B. (2023) Resurrection of the genus Subulo Smith, 1827 for the gray brocket deer, with designation of a neotype. Journal of Mammalogy, 104, 619–633. https://doi.org/10.1093/jmammal/gyac068
- Bickham, J.W., Wood, C.C. & Patton, J.C. (1995) Biogeographic implications of Cytochrome b sequences and allozymes in Sockeye (Oncorhynchus nerka). Journal of Heredity, 86, 140–144. https://doi.org/10.1093/oxfordjournals.jhered.a111544
- Bickham, J.W., Patton, J.C., Schlitter, D.A., Rautenbach, I.L. & Honeycutt, R.L. (2004) Molecular phylogenetics, karyotypic diversity, and partition of the genus Myotis (Chiroptera: Vespertilionidae). Molecular Phylogenetics and Evolution, 33, 333–338. https://doi.org/10.1016/j.ympev.2004.06.012
- Bradley, R.D. & Baker, R.J. (2001) A test of the Genetic Species Concept: cytochrome‐b sequences and mammals. Journal of Mammalogy, 82, 960–973. https://doi.org/10.1644/1545-1542(2001)082%3C0960:ATOTGS%3E2.0.CO;2
- Cifuentes‐Rincón, A., Morales‐Donoso, J.A., Sandoval, E.D.P., Tomazella, I.M., Mantellatto, A.M.B., de Thoisy, B. & Duarte, J.M.B. (2020) Designation of a neotype for Mazama americana (Artiodactyla, Cervidae) reveals a cryptic new complex of brocket deer species. ZooKeys, 958, 143–164. https://doi.org/10.3897/zookeys.958.50300
- Clement, M., Posada, D. & Crandall, K.A. (2000) TCS: a computer program to estimate gene genealogies. Molecular Ecology, 9, 1657–1659. https://doi.org/10.1046/j.1365-294x.2000.01020.x
- De Lima, A., Torres Martínez, M.M., Duarte, J.M.B. & González, S. (2024) Craniometric differentiation suggests disruptive selection on body size among sympatric brocket deer. Hystrix, the Italian Journal of Mammalogy. Available from: https://www.researchgate.net/publication/388994186_Craniometric_differentiation_suggests_disruptive_selection_on_body_size_among_sympatric_brocket_deer (accessed 22 September 2025)
- Duarte, J.M.B., González, S. & Maldonado, J.E. (2008) The surprising evolutionary history of South American deer. Molecular Phylogenetics and Evolution, 49, 17–22. https://doi.org/10.1016/j.ympev.2008.07.009
- Escobedo‐Morales, L.A., Mandujano, S., Eguiarte, L.E., Rodríguez Rodríguez, M.A. & Maldonado, J.E. (2016) First phylogenetic analysis of Mesoamerican brocket deer Mazama pandora and Mazama temama (Cetartiodactyla: Cervidae) based on mitochondrial sequences: implications on Neotropical deer evolution. Mammalian Biology, 81, 303–313. https://doi.org/10.1016/j.mambio.2016.02.003
- Escobedo‐Morales, L.A., Castañeda‐Rico, S., Mandujano, S., León‐Paniagua, L. & Maldonado, J.E. (2023) First description of the mitochondrial genomes of the Central American brocket deer Mazama temama (Kerr, 1792) and the Yucatán Peninsula brocket deer Odocoileus pandora Merriam, 1901. Molecular Biology Reports, 50, 4851–4863. https://doi.org/10.1007/s11033-023-08407-3
- Escobedo‐Morales, L.A., Castañeda‐Rico, S., Mandujano, S., León‐Paniagua, L., Martínez‐Meyer, E. & Maldonado, J.E. (2025) Mitochondrial genome and ultraconserved elements reveal a species complex within the Central American brocket deer Mazama temama (Artiodactyla: Cervidae). Molecular Phylogenetics and Evolution, 211, 108400. https://doi.org/10.1016/j.ympev.2025.108400
- Feng, X., Park, D.S., Liang, Y., Pandey, R. & Papeş, M. (2019) Collinearity in ecological niche modeling: confusions and challenges. Ecology and Evolution, 9 (18), 10365–10376. https://doi.org/10.1002/ece3.5555
- Fick, S.E. & Hijmans, R.J. (2017) WorldClim 2: new 1‐km spatial resolution climate surfaces for global land areas. International Journal of Climatology, 37 (12), 4302–4315. https://doi.org/10.1002/joc.5086
- Gong, L., Shi, W., Si, L.Z., Wang, Z.M. & Kong, X.Y. (2015) The complete mitochondrial genome of peacock sole Pardachirus pavoninus (Pleuronectiformes: Soleidae) and comparative analysis of the control region among 13 soles. Molecular Biology, 49, 408–417. https://doi.org/10.1134/S0026893315030061
- Groves, C. & Grubb, P. (2011) Ungulate taxonomy. John Hopkins University Press, Baltimore, Maryland, 317 pp. https://doi.org/10.56021/9781421400938
- Gutiérrez, E.E., Maldonado, J.E., Radosavljevic, A., Molinari, J., Patterson, B.D., Martínez‐C, J.M., Rutter, A.R., Hawkins, M.T.R., Garcia, F.J. & Helgen, K.M. (2015) The taxonomic status of Mazama bricenii and the significance of the Táchira Depression for mammalian endemism in the Cordillera de Mérida, Venezuela. PLoS ONE, 10, 1–24. https://doi.org/10.1371/journal.pone.0129113
- Gutiérrez, E.E., Helgen, K.M., McDonough, M.M., Bauer, F., Hawkins, M.T.R., Escobedo‐Morales, L.A., Patterson, B.D. & Maldonado, J.E. (2017) A gene‐tree test of the traditional taxonomy of American deer: the importance of voucher specimens, geographic data, and dense sampling. ZooKeys, 697, 87–131. https://doi.org/10.3897/zookeys.697.15124
- Hall, T.A. (1999) BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
- Heckeberg, N.S. (2020) The systematics of the Cervidae: a total evidence approach. PeerJ, 8, e8114. https://doi.org/10.7717/peerj.8114
- Heckeberg, N.S., Erpenbeck, D., Wörheide, G. & Rössner, G.E. (2016) Systematic relationships of five newly sequenced cervid species. PeerJ, 4, e2307. https://doi.org/10.7717/peerj.2307
- Hershkovitz, P. (1982) Neotropical deer (Cervidae): part 1. Pudus, genus Pudu Gray. Fieldiana Zoology, 11, 1–86. https://doi.org/10.5962/bhl.title.5080
- Hoang, D.T., Chernomor, O., Von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2018) UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution, 35, 518–522. https://doi.org/10.1093/molbev/msx281
- IUCN Standards and Petitions Subcommittee (2010) Guidelines for using the IUCN Red List Categories and Criteria. Version 8.1. IUCN, Gland, Switzerland, 114 pp.
- Jasper, J.G., Lee, Jr. T.E., Zabel, C.J., Twohy, C.L., Lane, K.K. & Robertson, C.S. (2022) Mazama rufina (Artiodactyla: Cervidae). Mammalian Species, 54 (1016), 212–219. https://doi.org/10.1093/mspecies/seac001
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587–589. https://doi.org/10.1038/nmeth.4285
- Kass, J.M., Pinilla-Buitrago, G.E., Paz, A., Johnson, B.A., Grisales-Betancur, V., Meenan, S.I., Attali, D., Broennimann, O., Galante, P.J., Maitner, B.S., Owens, H.L., Varela, S., Aiello-Lammens, M.E., Merow, C., Blair, M.E. & Anderson, R.P. (2023) Wallace 2: a shiny app for modeling species niches and distributions redesigned to facilitate expansion via module contributions. Ecography, 2023 (3), e06547, 1–9. https://doi.org/10.1111/ecog.06547
- Kass, J.M., Vilela, B., Aiello‐Lammens, M.E., Muscarella, R., Merow, C. & Anderson, R.P. (2018) Wallace: A flexible platform for reproducible modeling of species niches and distributions built for community expansion. Methods in Ecology and Evolution, 9, 1151–1156. https://doi.org/10.1111/2041-210X.12945
- Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P. & Drummond, A. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28 (12), 1647–1649. https://doi.org/10.1093/bioinformatics/bts199
- Leigh, J.W. & Bryant, D. (2015) POPART: full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6, 1110–1116. https://doi.org/10.1111/2041-210X.12410
- Lizcano, D.J. (2006) Ecology and conservation of large mammals in the northern Andes. Ph.D. dissertation, University of Kent, Kent, Canterbury. [unknown pagination]
- Lizcano, D. & Alvarez, S.J. (2016) Mazama rufina. The IUCN Red List of Threatened Species, 2016, e.T12914A22165586. Available from: https://doi.org/ 10.2305/IUCN.UK.2016-2.RLTS.T12914A22165586.en (accessed 3 November 2022)
- Mammal Diversity Database (2025) Mammal Diversity Database. Version 2.0. Data Set. Zenodo. Available from: https://zenodo.org/records/15007505 (accessed 22 September 2025) https://doi.org/10.5281/zenodo.15007505
- Minh, B.Q., Nguyen, M.A. & von Haeseler, A. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution, 30, 1188–1195. https://doi.org/10.1093/molbev/mst024
- Morales-Donoso, J.A., Vacari, G.Q., Bernegossi, A.M., Sandoval, E.D.P., Peres, P.H.F., Galindo, D.J., de Thoisy, B., Vozdova, M., Kubickova, S. & Duarte, J.M.B. (2023) Revalidation of Passalites Gloger, 1841 for the Amazon brown brocket deer P. nemorivagus (Cuvier, 1817) (Mammalia, Artiodactyla, Cervidae). ZooKeys, 1167, 241–264. https://doi.org/10.3897/zookeys.1167.100577
- Muscarella, R., Galante, P.J., Soley-Guardia, M., Boria, R.A., Kass, J.M., Uriarte, M. & Anderson, R.P. (2014) ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods in Ecology and Evolution, 5 (11), 1198–1205. https://doi.org/10.1111/2041-210X.12261
- Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274. https://doi.org/10.1093/molbev/msu300
- Pearson, R.G., Raxworthy, C.J., Nakamura, M. & Peterson, T.A. (2007) Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. Journal of Biogeography, 34 (1), 102–117. https://doi.org/10.1111/j.1365-2699.2006.01594.x
- Peres, P.H.F., Luduvério, D.J., Bernegossi, A.M., Galindo, D.J., Nascimento, G.B., Oliveira, M.L., Sandoval, E.D.P., Vozdova, M., Kubickova, S. & Duarte, J.M.B. (2021) Revalidation of Mazama rufa (Illiger 1815) (Artiodactyla: Cervidae) as a Distinct Species out of the Complex Mazama americana (Erxleben 1777). Frontiers in Genetics, 12, 742870. https://doi.org/10.3389/fgene.2021.742870
- Phillips, S.J., Anderson, R.P., Dudík, M., Schapire, R.E. & Blair, M.E. (2017) Opening the black box: an open-source release of Maxent. Ecography, 40 (7), 887–893. https://doi.org/10.1111/ecog.03049
- Phillips, S.J. & Dudík, M. (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography, 31 (2), 161–288. https://doi.org/10.1111/j.0906-7590.2007.5203.x
- Ramírez-Chaves, H.E., Ossa-López, P.A., Lasso-Lasso, L., Rivera-Páez, F.A., Roncancio Duque, N. & Escobedo-Morales, L.A. (2021) Range extension of the Central American Red Brocket, Mazama temama (Kerr, 1792) (Artiodactyla, Cervidae) in Colombia. Check List, 17, 1095–1102. https://doi.org/10.15560/17.4.1095
- Sandoval, E.D.P., Rola, L.D., Morales-Donoso, J.A., Gallina, S., Reyna-Hurtado, R. & Duarte, J.M.B. (2022) Integrative analysis of Mazama temama (Artiodactyla: Cervidae) and designation of a neotype for the species. Journal of Mammalogy, 103, 447–458. https://doi.org/10.1093/jmammal/gyab169
- Sandoval, E.D.P., Jędrzejewski, W., Molinari, J., Vozdova, M., Cernohorska, H., Kubickova, S., Bernegossi, A.M., Caparroz, R. & Duarte, J.M.B. (2024) Description of Bisbalus, a new genus for the gray brocket, Mazama cita Osgood, 1912 (Mammalia, Cervidae), as a step to solve the Neotropical deer puzzle. Taxonomy, 4, 10–26. https://doi.org/10.3390/taxonomy4010002
- Shcheglovitova, M. & Anderson, R.P. (2013) Estimating optimal complexity for ecological niche models: a jackknife approach for species with small sample sizes. Ecological Modelling, 269, 9–17. https://doi.org/10.1016/j.ecolmodel.2013.08.011
- Solari, S., Muñoz-Saba, Y., Rodríguez-Mahecha, J.V., Defler, T.R., Ramírez-Chaves, H.E. & Trujillo, F. (2013) Riqueza, endemismo y conservación de los mamíferos de Colombia. Mastozoología Neotropical, 20, 301–365.
- Tamura, K., Stecher, G. & Kumar, S. (2021) MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution, 38 (7), 3022–3027. https://doi.org/10.1093/molbev/msab120
- Thomas, O. (1908) A new deer of the brocket group from Venezuela. Annals and Magazine of Natural History, Series 8, 1 (4), 349–350. https://doi.org/10.1080/00222930808692415
- Thomas, O. (1913) On certain of the smaller S.-American Cervidae. Annals and Magazine of Natural History, Series 8, 11 (66), 585–589. https://doi.org/10.1080/00222931308693360
- von den Driesch, A. (1976) A guide to the measurement of animal bones from archaeological sites. Harvard University Press, Peabody Museum of Archaeology and Ethnology, Cambridge, Massachusetts, 136 pp.