Abstract
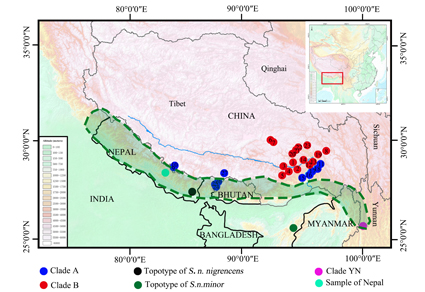
The Himalayan shrew, Soriculus nigrescens Gray, 1842, belongs to the monotypic genus Soriculus, which is distributed mainly in the Himalayan region. Previous authors have studied its classification based on morphological and molecular data. However, no comprehensive study of the diversity and phylogeny of this species has been performed. In this study, we investigated the molecular phylogeny, genetic diversity, and species divergence of S. nigrescens based on one mitochondrial gene and three nuclear genes. A total of 124 samples from 27 sites in Southwest China were analyzed. Our molecular phylogenetic analyses and species divergence reveal non-monophyly of Soriculus, potentially representing two genera and three clades. Populations from Yunnan (Clade YN) represent the subspecies S. n. minors and should recover the full species status. Populations from Himalayas (Clade A) represent the species S. nigrescens, while populations from southeastern Nyenchen Tanglha Mountains and southern Himalayas (Clade B) represent a new cryptic and unnamed species. Species delimitation analyses and deep genetic distance analysis clearly support the species status of these three evolving clades. The putative new genus and cryptic species should be studied and identified in the future using a more extensive sampling combined with a comprehensive morphological and phylogenetic analysis.
References
- Burgin, C.J. & He, K. (2018) Family Soricidae (shrew) In: Wilson, D.E. & Russell, A.M. (Eds.), Hand book of the mammals of the world: insectivores, sloths and colugos. Barcelona: Lynx Edicions, pp. 474–530.
- Chen, S., Liu, S., Liu, Y., Kai, H., Chen, W. & Zhang, X. (2012) Molecular phylogeny of Asiatic short- tailed shrews, genus Blarinella Thomas, 1911 (Mammalia: Soricomorpha: Soricidae) and its taxonomic implications. Zootaxa, 3250 (1), 43–53. https://doi.org/10.11646/zootaxa.3250.1.3
- Chen, S., Sun, Z., He, K., Jiang, X., Liu, Y., Koju, N.P., Zhang, X., Tu, F., Fan, Z., Liu, S. & Yue, B. (2015) Molecular phylogenetics and phylogeographic structure of Sorex bedfordiae based on mitochondrial and nuclear DNA sequences. Molecular Phylogenetics Evolution, 84, 245–253. https://doi.org/ 10.1016/j.ympev.2014.12.016
- Chen, S., Jiao, Q., Liu, Z., Liu, Y. & Liu, S. (2020) Multilocus phylogeny and cryptic diversity of white-toothed shrews (Mammalia, Eulipotyphla, Crocidura) in China. BMC Evolutionary Biology, 20, 29. https://doi.org/10.1186/s12862-020-1588-8
- Corbett, G.B. & Hill, J.E. (1992) The mammals of the Indomalay region: A systematic review. Natural History Museum Publications, Oxford University Press, vii 488 pages. ISBN 0-19-854693-9. Price: £60.00 (hardback) Journal of Tropical Ecology, 9 (3), 338. https://doi.org/10.1017/S0266467400007380
- Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) JModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772–772. https://doi.org/ 10.1038/nmeth.2109
- Ding, W.N., Ree, R.H., Spicer, R.A. & Xing, Y.W. (2020) Ancient orogenic and monsoon-driven assembly of the world's richest temperate alpine flora. Science, 369 (6503), 578–581. https://doi.org/ 10.1126/science.abb4484
- Drummond, A.J. & Rambaut, A. (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7 (1), 214. https://doi.org/ 10.1186/1471-2148-7-214
- Drummond, A.J., Suchard, M.A., Dong, X. & Rambaut, A. (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 29 (8), 1969–1973. https://doi.org/ 10.1093/molbev/mss075
- Ellerman, J.R. & Morrison-Scott, T.C.S. (1951) Checklist of Palaearctic and Indian Mammals1758-1946. London: British Museum (Natural History).
- Evanno, G.S., Regnaut, S.J. & Goudet, J. (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology, 14 (8), 2611–2620. https://doi.org/ 10.1111/j.1365-294X.2005. 02553. x
- Feijó, A., Ge, D.Y., Wen, Z.X., Cheng, J.L., Xia, L., Patterson, B.D. & Yang, Q.S. (2022) Mammalian diversification bursts and biotic turnovers are synchronous with Cenozoic geoclimatic events in Asia. Proceedings of the National Academy of Sciences, 119, 49. https://doi.rorg//10.1073/pnas.2207845119
- He, K., Li, Y.J., Brandley, M.C., Lin, L.K. & Jiang, X.L. (2010) A multi-locus phylogeny of Nectogalini shrews and influences of the paleoclimate on speciation and evolution. Molecular Phylogenetics & Evolution, 56 (2), 734–746. https://doi.org/ 10.1016/j.ympev.2010.03.039
- He, K., Chen, X., Chen, P., He, S.W., Cheng, F., Jiang, X.L. & Campbell, K. (2018) A new genus of Asiatic short-tailed shrew (Soricidae, Eulipotyphla) based on molecular and morphological comparisons. Zoological Research. 39 (5), 321–334. https://doi.org/ 10.24272/j.issn.2095-8137.2018.058
- Heled, Joseph. & Alexei, J.Drummond. (2008) Bayesian inference of population size history from multiple loci. BMC Evolutionary Biology, 8, 289–289. https://doi.org/ 10.1186/1471-2148-8-289
- Hinton, M.A.C. (1922) Scientific results from the mammal survey. No. 34. The house rats of Nepal. Journal of the Bombay Natural History Society, 28, 1056–1066.
- Hoffmann, R.S. (1986) A review of the genus Soriculus (Mammalia: Insectivora). Journal of the Bombay Natural History Society, 82, 459–481.
- Hoffmann, R.S. (1987) A review of the systematics and distribution of Chinese red-toothed shrews (mammalia: soricinae). Acta Theriologica Sinica, 7 (2), 100–139.
- Huelsenbeck, J. & Rannala, B. (2004) Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Systematic Biology, 53 (6), 904–13. https://doi.org/ 10.1080/10635150490522629
- Hutterer, R. (1993) Order Insectivora. Mammal Species of the World, 1993, 69–130.
- Hutterer, R. (2005) Order Soricomorpha. In: Wilson, D.E. & Reeder, D.M. (Eds.), Mammal Species of the World 3rd ed. The Johns Hopkins University Press, Baltimore, pp. 220–311.
- Librado, P. & Rozas, J. (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. https://doi.org/ 10.1093/bioinformatics/btp187
- Motokawa, M. (2003) Soriculus minor Dobson, 1890, senior synonym of S. radulus Thomas, 1922 (Insectivora, Soricidae). Mammalian Biology, 68 (3), 178–180. https://doi.org/ 10.1078/1616-5047-00080
- Motokawa, M., Harada, M., Mekada, K. & Shrestha, K.C. (2008) Karyotypes of three shrew species (Soriculus nigrescens, Episoriculus caudatus and Episoriculus sacratus) from Nepal. Integrative Zoology, 3 (3), 180–185. https://doi.org/10.1111/j.1749-4877.2008.00097.x
- Ohdachi, S.D., Hasegawa, M., Iwasa, M.A., Vogel, P., Oshida, T. & Lin, L.K. (2006) Molecular phylogenetics of soricid shrews (Mammalia) based on mitochondrial cytochrome b gene sequences: with special reference to the Soricinae. Proceedings of the Zoological Society of London, 270 (1), 177–191.
- https://doi.org/ 10.1111/j.1469-7998.2006.00125.x
- Pritchard, J.K., Stephens, M. & Donnelly, P. (2000) Inference of population structure using multilocus genotype data. Genetics. Jun,155 (2), 945–959. https://doi.org/ 10.1093/genetics/155.2.945
- Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England), Aug, 19 (12), 1572–1574. https://doi.org/ 10.1093/bioinformatics/btg180
- Sambrook, J. & Russell, D.W. (2006) Molecular Cloning, 4th ed. Cold Spring Harbor Laboratory Press, New York. https://doi.org/10.1101/pdb.prot3919
- Smith, A.T. & Xie, Yan. (2009) A Guide to the Mammals of China. Princeton University Press. Princeton, New Jersey.
- Spicer, R.A. (2017,) Tibet, the Himalaya, Asian monsoons and biodiversity- In what ways are they related. Plant Diversity, 2017, 39 (5):12, 233–244. https://doi.org/10.1016/j.pld.2017.09.001
- Springer, M.S., Murphy, W.J. & Roca, A.L. (2018). Appropriate fossil calibrations and tree constraints uphold the Mesozoic divergence of solenodons from other extant mammals. Molecular Phylogenetics & Evolution, S1055790317300994. https://doi.org/10.1016/j.ympev.2018.01.007
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. & Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739. https://doi.org/ 10.1093/molbev/msr121
- Trifinopoulos, Jana., Nguyen, Lam-Tung., von, Haeseler, Arndt. & Minh, B.Q. (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic acids research. 8, 44 (W1), W232–5. https://doi.org/10.1093/nar/gkw256
- Xu, W., Dong, W.J., Fu, T.T., Gao, W., Lu, C.Q., Yan. F., Wu, Y.H. Jiang, K., Jin, J.Q. Chen, H.M., Zhang, Y.P., Hillis, D.M. & Che, J. (2021) Herpetological phylogeographic analyses support a Miocene focal point of Himalayan uplift and biological diversification, National Science Review, 8, 9, nwaa263. https://doi.org/10.1093/nsr/nwaa263
- Yang, Z. & Rannala, B. (2014) Unguided species delimitation using DNA sequence data from multiple Loci. Molecular Biology and Evolution, 31, 3125–3135. https://doi.org/ 10.1093/molbev/msu279