Abstract
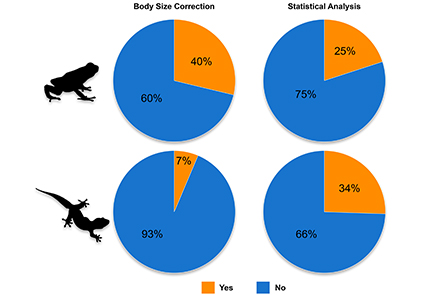
Although body size correction and inferential statistics have been used in morphological studies for many decades, their applications are far from being ubiquitous. We performed a meta-analysis to quantify the extent of taxonomic papers that performed body size correction and implemented a statistical hypothesis testing framework during the analysis of morphological data. Our results indicate that in most papers, neither of these analyses were performed but instead, cursory comparisons of descriptive statistics were presented. With the development of numerous freely available and powerful statistical programs such as R, we find it prudent to outline a standardized and statistically defensible framework to enhance the workflow of morphological analyses in taxonomic studies. This 5-step approach can be applied to meristic and mensural data across a wide range of taxonomic groups. We include an easy-to-use companion R script to facilitate the implementation of this workflow. Our proposed framework is not rooted in phylogenetic or evolutionary theory and hence, should not be used in place of explicit species delimitation techniques. Nevertheless, it can be incorporated into a more robust integrative taxonomic framework and is particularly useful for identifying diagnostic characters for species diagnoses.
References
Blanca, M.J., Alarcón, R., Arnau, J., Bono, R. & Bendayan, R. (2017) Non-normal data: Is ANOVA still a valid option? Psicothema, 29, 552–557. https://doi.org/10.7334/psicothema2016.383
Blanca, M.J., Alarcón, R., Arnau, J., Bono, R. & Bendayan, R. (2018) Effect of variance ratio on ANOVA robustness: Might 1.5 be the limit? Behavior Research Methods, 50, 937–962. https://doi.org/10.3758/s13428-017-0918-2
Bolnick, D.I., Amarasekare, P., Araújo, M.S., Bürger, R., Levine, J.M., Novak, M., Rudolf, V.H.W., Schreiber, S.J., Urban, M.C. & Vasseur, D.A. (2011) Why intraspecific trait variation matters in community ecology. Trends in Ecology and Evolution, 26, 183–192. https://doi.org/10.1016/j.tree.2011.01.009
Cardinal, L.J. (2015) Central tendency and variability in biological systems: Part 2. Journal of Community Hospital Internal Medicine Perspectives, 5, 28972. https://doi.org/10.3402/jchimp.v5.28972
Chan, K.O. & Grismer, L.L. (2021) Correcting for body size variation in morphometric analysis. bioRxiv, 2021.05.17.444580. https://doi.org/10.1101/2021.05.17.444580
Halsey, L.G. (2019) The reign of the p-value is over: What alternative analyses could we employ to fill the power vacuum? Biology Letters, 15, 20190174. https://doi.org/10.1098/rsbl.2019.0174
Klauber, L.M. (1943) The correlation of variability within and between Rattlesnake populations. Copeia, 2, 115–118. https://doi.org/10.2307/1437777
Lleonart, J., Salat, J. & Torres, G.J. (2000) Removing allometric effects of body size in morphological analysis. Journal of Theoretical Biology, 205, 85–93. https://doi.org/10.1006/jtbi.2000.2043
Padial, J.M., Miralles, A., De la Riva, I. & Vences, M. (2010) The integrative future of taxonomy. Frontiers in Zoology, 7, 16. https://doi.org/10.1186/1742-9994-7-16
R Core Team (2014) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. [program]
Reist, J.D. (1985) An empirical evaluation of several univariate methods that adjust for size variation in morphometric data. Canadian Journal of Zoology, 63, 1429–1439. https://doi.org/10.1139/z85-213
Rivera-correa, M., Baldo, D., Candioti, F.V., Goyannes, V., Orrico, D., Blackburn, D.C., Castroviejo-Fisher, S., Chan, K.O., Gambale, P., Gower, D.J., Quah, E.S.H., Rowley, J.J.L., Twomey, E. & Vences, M. (2021) Amphibians in Zootaxa: 20 years documenting the global diversity of frogs, salamanders, and caecilians. Zootaxa, 4979 (1), 57–69. https://doi.org/10.11646/zootaxa.4979.1.9
Schmider, E., Ziegler, M., Danay, E., Beyer, L. & Bühner, M. (2010) Is It Really Robust?: Reinvestigating the robustness of ANOVA against violations of the normal distribution assumption. Methodology, 6, 147–151. https://doi.org/10.1027/1614-2241/a000016
Senthamarai Kannan, K. & Manoj, K. (2015) Outlier detection in multivariate data. Applied Mathematical Sciences, 9, 2317–2324. https://doi.org/10.12988/ams.2015.53213
Thorpe, R.S. (1975) Quantitative handling of characters useful in snake systematics with particular reference to intraspecific variation in the Ringed Snake Natrix natrix. Biological Journal of the Linnean Society, 7, 27–43. https://doi.org/10.1111/j.1095-8312.1975.tb00732.x
Thorpe, R.S. (1983) A review of the numerical methods for recognising and analysing racial differentiation. In: Felsenstein, J. (Ed.), Numerical Taxonomy. Springer Verlag, Berlin and Heidelberg, pp. 404–423. https://doi.org/10.1007/978-3-642-69024-2_43
Wasserstein, R.L., Schirm, A.L. & Lazar, N.A. (2019) Moving to a World Beyond “p < 0.05.” American Statistician, 73, 1–19. https://doi.org/10.1080/00031305.2019.1583913