Abstract
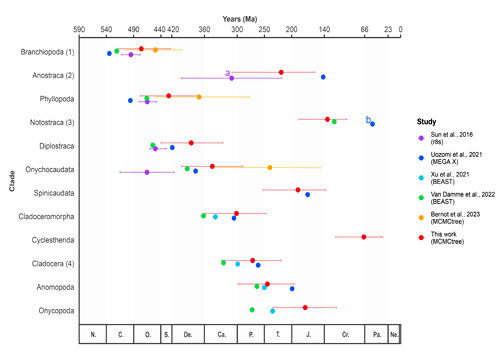
An understanding of Branchiopoda’s evolutionary history is crucial for a comprehensive knowledge of the Pancrustacea tree of life, given their close evolutionary relationship with Hexapoda. Despite significant advances in molecular and morphological phylogenetics that have resolved much of the branchiopod backbone topology, a reliable temporal framework remains elusive. Key challenges include a sparse fossil record, long-term morphological stasis, and past topological inconsistencies. Leveraging a Bayesian Inference approach and the most extensive phylogenomic dataset for branchiopod to date, encompassing 46 species and over 130 genes, we inferred a time-calibrated phylogenetic tree. Furthermore, to strengthen the confidence in our divergence times estimation, we assessed the impact of age priors, topological uncertainties, and gene trees which are discordant from the species trees. Our results are largely consistent with the fossil record and with previous studies, indicating that Branchiopoda originated between 400 and 500 million years ago, and the orders of large branchiopods diversified during the Mesozoic. Concerning Cladocera, results remain problematic, with a sharper uncertainty in the diversification time with respect to the fossil record. Though, the jackknife resampling of fossils and the other sensitivity analyses proved our calibration method to be robust, suggesting that the difficulties in obtaining a paleontological-consistent time tree may be hindered by the variability in branchiopod substitution rates and topological instability within certain clades.
References
- Abascal, F., Zardoya, R. & Telford, M.J. (2010) Translatorx: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Research, 38 (Suppl 2), W7–W13. https://doi.org/10.1093/nar/gkq291
- Alencar, L.R., Quental, T.B., Grazziotin, F.G., Alfaro, M.L., Martins, M., Venzon, M. & Zaher, H. (2016) Diversification in vipers: Phylogenetic relationships, time of divergence and shifts in speciation rates. Molecular Phylogenetics and Evolution, 105, 50–62. https://doi.org/10.1016/j.ympev.2016.07.029
- Ax, P. (1999) Das System der Metazoa II. Ein Lehrbuch der phylogenetischen Systematik. Fischer, Stuttgart, 384 pp.
- Baldwin-Brown, J.G., Weeks, S.C. & Long, A.D. (2018) A new standard for crustacean genomes: the highly contiguous, annotated genome assembly of the clam shrimp Eulimnadia texana reveals HOX gene order and identifies the sex chromosome. Genome Biology and Evolution, 10 (1), 143–156. https://doi.org/10.1093/gbe/evx280
- Bánki, O., Roskov, Y., Döring, M., Ower, G., Hernández Robles, D.R., Plata Corredor, C.A., Stjernegaard Jeppesen, T., Örn, A., Vandepitte, L., Hobern, D., Schalk, P., DeWalt, R.E., Ma, K., Miller, J., Orrell, T., Aalbu, R., Abbott, J., Adlard, R., Aedo, C., et al. (2024) Catalogue of Life (Annual Checklist 2024). Catalogue of Life, Amsterdam, Netherlands. https://doi.org/10.48580/dgmv5
- Bernot, J.P., Owen, C.L., Wolfe, J.M., Meland, K., Olesen, J. & Crandall, K.A. (2023) Major revisions in pancrustacean phylogeny and evidence of sensitivity to taxon sampling. Molecular Biology and Evolution, 40 (8), msad175. https://doi.org/10.1007/978-1-4020-8259-7_18
- Brendonck, L., Rogers, D.C., Olesen, J., Weeks, S. & Hoeh, W.R. (2008) Global diversity of large branchiopods (Crustacea: Branchiopoda) in freshwater. Freshwater Animal Diversity Assessment, 595, 167–176.
- Capella-Gutiérrez, S., Silla-Martıénez, J.M. & Gabaldòn, T. (2009) Trimal: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 25 (15), 1972–1973. https://doi.org/10.1093/bioinformatics/btp348
- Carruthers, T., Sun, M., Baker, W. J., Smith, S.A., De Vos, J.M. & Eiserhardt, W.L. (2022) The implications of incongruence between gene tree and species tree topologies for divergence time estimation. Systematic Biology, 71 (5), 1124–1146. https://doi.org/10.1093/sysbio/syac012
- Castellucci, F., Luchetti, A. & Mantovani, B. (2022) Exploring mitogenome evolution in Branchiopoda (Crustacea) lineages reveals gene order rearrangements in Cladocera. Scientific Reports, 12 (1), 4931. https://doi.org/10.1038/s41598-022-08873-y
- Chen, P.J. & Shen, Y.B. (1985) An introduction to fossil Conchostraca. Science Press, Beijing, 241 pp. [In Chinese]
- Colbourne, J.K., Pfrender, M.E., Gilbert, D., Thomas, W.K., Tucker, A., Oakley, T.H., Tokishita, S., Aerts, A., Arnold, G.J., Basu, M.K., Bauer, D.J., Cáceres, C.E., Carmel, L., Casola, C., Choi, J.-H., Detter, J.C., Dong, Q., Dusheyko, S., Eads, B.D., Fröhlich, T., Geiler-Samerotte, K.A., Gerlach, D., Hatcher, P., Jogdeo, S., Krijgsveld, J., Kriventseva, E.V., Kültz, D., Laforsch, C., Lindquist, E., Lopez, J., Manak, J.R., Muller, J., Pangilinan, J., Patwardhan, R.P., Pitluck, S., Pritham, E.J., Rechtsteiner, A., Rho, M., Rogozin, I.B., Sakarya, O., Salamov, A., Schaack, S., Shapiro, H., Shiga, Y., Skalitzky, C., Smith, Z., Souvorov, A., Sung, W., Tang, Z., Tsuchiya, D., Tu, H., Vos, H., Wang, M., Wolf, Y.I., Yamagata, H., Yamada, T., Ye, Y., Shaw, J.R., Andrews, J., Crease, T.J., Tang, H., Lucas, S.M., Robertson, H.M., Bork, P., Koonin, E.V., Zdobnov, E.M., Grigoriev, I.V., Lynch, M. & Boore, J.L. (2011) The ecoresponsive genome of Daphnia pulex. Science, 331 (6017), 555–561. https://doi.org/10.1126/science.1197761
- Forni, G., Martelossi, J., Valero, P., Hennemann, F.H., Conle, O., Luchetti, A. & Mantovani, B. (2022) Macroevolutionary analyses provide new evidence of phasmid wings evolution as a reversible process. Systematic Biology, 71 (6), 1471–1486. https://doi.org/10.1093/sysbio/syac038
- Forro, L., Korovchinsky, N.M., Kotov, A.A. & Petrusek, A. (2008) Global diversity of cladocerans (Cladocera; Crustacea) in freshwater. Freshwater Animal Diversity Assessment, 595, 177–184. https://doi.org/10.1007/978-1-4020-8259-7_19
- Fyer, G. (1987) A new classification of the branchiopod Crustacea. Zoological Journal of the Linnean Society, 91(4), 357–383. https://doi.org/10.1111/j.1096-3642.1987.tb01420.x
- Fu, L.M., Niu, B.F., Zhu, Z.W., Wu, S.T. & Li, W.Z. (2012). CD-HIT: Accelerated for clustering the nextgeneration sequencing data. Bioinformatics, 28 (23), 3150–3152. https://doi.org/10.1093/bioinformatics/bts565
- Grabherr, M.G., Haas, B.J., Yassour, M., Levin, J.Z., Thompson, D.A., Amit, I., Adiconis, X., Fan, L., Raychowdhury, R., Zeng, Q.D., Chen, Z.H., Mauceli, E., Hacohen, N., Gnirke, A., Rhind, N., di Palma, F., Birren, B.W., Nusbaum, C., Lindblad-Toh, K., Friedman, N. & Regev, A. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 29 (7), 644–652. https://doi.org/10.1038/nbt.1883
- Grau-Bové, X., Navarrete, C., Chiva, C., Pribasnig, T., Antó, M., Torruella, G., Galindo, L. J., Lang, B.F., Moreira, D., López-Garcia, P., Ruiz-Trillo, I., Schleper, C., Sabidó, E. & Sebé-Pedrós, A. (2022) A phylogenetic and proteomic reconstruction of eukaryotic chromatin evolution. Nature Ecology & Evolution, 6 (7), 1007–1023. https://doi.org/10.1038/s41559-022-01771-6
- Gueriau, P., Rabet, N., Clément, G., Lagebro, L., Vannier, J., Briggs, D.E.G., Charbonnier, S., Olive, S. & Béthoux, O. (2016) A 365-million-year-old freshwater community reveals morphological and ecological stasis in branchiopod crustaceans. Current Biology, 26 (3), 383–390. https://doi.org/10.1016/j.cub.2015.12.039
- Hegna, T.A. (2012) Phylogeny and fossil record of branchiopod crustaceans: An integrative approach. Unpublished Ph.D. thesis, Yale University, New Haven, CT.
- Hegna, T.A. & Astrop, T.I. (2020) The fossil record of the clam shrimp (Crustacea; Branchiopoda). Zoological Studies, 59, 43. https://doi.org/10.6620/zs.2020.59-43
- Huang, D.Y., Cai, C.Y., Fu, Y.Z. & Su, Y.T. (2018). The Middle-Late Jurassic Yanliao entomofauna. Palaeoentomology, 1 (1), 3–31. https://doi.org/10.11646/palaeoentomology.1.1.2
- Ikeda, K.T., Hirose, Y., Hiraoka, K., Noro, E., Fujishima, K., Tomita, M. & Kanai, A. (2015) Identification, expression, and molecular evolution of microRNAs in the “living fossil” Triops cancriformis (tadpole shrimp). RNA, 21 (2), 230–242. https://doi.org/10.1261/rna.045799.114
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14 (6), 587–589. https://doi.org/10.1038/nmeth.4285
- Kieran Blair, S.R., Hull, J., Escalona, M., Finger, A., Joslin, S.E.K., Sahasrabudhe, R., Marimuthu, M.P.A., Nguyen, O., Chumchim, N., Reister Morris, E., Velazquez, S. & Schreier, A. (2022) The reference genome of the Vernal Pool Tadpole Shrimp, Lepidurus packardi. Journal of Heredity, 113 (6), 706–711. https://doi.org/10.1093/jhered/esac051
- Kieran Blair, S.R., Schreier, A., Escalona, M., Finger, A.J., Joslin, S.E.K., Sahasrabudhe, R., Marimuthu, M.P.A., Nguyen, O., Chumchim, N., Reister Morris, E., Mangelson, H. & Hull, J. (2023a) A chromosome-level reference genome for the Versatile Fairy Shrimp, Branchinecta lindahli. Journal of Heredity, 114 (1), 74–80. https://doi.org/10.1093/jhered/esac057
- Kieran Blair, S.R., Schreier, A., Escalona, M., Finger, A.J., Joslin, S.E.K., Sahasrabudhe, R., Marimuthu, M.P.A., Nguyen, O., Chumchim, N., Reister Morris, E., Mangelson, H. & Hull, J. (2023b) A draft reference genome of the Vernal Pool Fairy Shrimp, Branchinecta lynchi. Journal of Heredity, 114 (1), 81–87. https://doi.org/10.1093/jhered/esac056
- Kotov, A. A. (2007). Jurassic Cladocera (Crustacea, Branchiopoda) with a description of an extinct Mesozoic order. Journal of Natural History, 41 (1-4), 13–37. https://doi.org/10.1080/00222930601164445
- Kotov, A. A. (2009a). A revision of the extinct Mesozoic family Prochydorusidae Smirnov, 1992 (Crustacea: Cladocera) with a discussion of its phylogenetic position. Zoological Journal of the Linnean Society, 155 (2), 253–265. https://doi.org/10.1111/j.1096-3642.2008.00412.x
- Kotov, A.A. (2009b) New finding of Mesozoic ephippia of the Anomopoda (Crustacea: Cladocera). Journal of Natural History, 43 (9-10), 523–528. https://doi.org/10.1080/00222930802003020
- Kotov, A.A. & Taylor, D.J. (2011) Mesozoic fossils (> 145 Mya) suggest the antiquity of the subgenera of Daphnia and their coevolution with chaoborid predators. BMC Evolutionary Biology, 11, 1–9. https://doi.org/10.1186/1471-2148-11-129
- Lee, B.Y., Choi, B.S., Kim, M.S., Park, J.C., Jeong, C.B., Han, J. & Lee, J.S. (2019) The genome of the freshwater water flea Daphnia magna: A potential use for freshwater molecular ecotoxicology. Aquatic Toxicology, 210, 69–84. https://doi.org/10.1016/j.aquatox.2019.02.009
- Lozano-Fernandez, J., Giacomelli, M., Fleming, J.F., Chen, A., Vinther, J., Thomsen, P. F., Glenner, H., Palero, F., Legg, D.A. Iliffe, T.M, Pisani, D. & Olesen, J. (2019) Pancrustacean evolution illuminated by taxon-rich genomic-scale data sets with an expanded remipede sampling. Genome Biology and Evolution, 11 (8), 2055–2070. https://doi.org/10.1093/gbe/evz097
- Luchetti, A., Forni, G., Martelossi, J., Savojardo, C., Martelli, P.L., Casadio, R., Skaist, A. M., Wheelan, S.J. & Mantovani, B. (2021) Comparative genomics of tadpole shrimps (Crustacea, Branchiopoda, Notostraca): Dynamic genome evolution against the backdrop of morphological stasis. Genomics, 113 (6), 4163–4172. https://doi.org/10.1016/j.ygeno.2021.11.001
- Mathers, T.C., Hammond, R.L., Jenner, R.A., Hänfling, B. & Gomez, A. (2013) Multiple global radiations in tadpole shrimps challenge the concept of ‘living fossils’. PeerJ, 1, e62. https://doi.org/10.7717/peerj.62
- Mendes, F.K. & Hahn, M.W. (2016) Gene tree discordance causes apparent substitution rate variation. Systematic Biology, 65 (4), 711–721. https://doi.org/10.1093/sysbio/syw018
- Nguyen, L.-T., Schmidt, H.A., Von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32 (1), 268–274. https://doi.org/10.1093/molbev/msu300
- Nickel, J., Schell, T., Holtzem, T., Thielsch, A., Dennis, S.R., Schlick-Steiner, B.C., Steiner, F.M., Möst, M., Pfenninger, M., Schwenk, K. & Cordellier, M. (2021) Hybridization dynamics and extensive introgression in the Daphnia longispina species complex: new insights from a high-quality Daphnia galeata reference genome. Genome Biology and Evolution, 13 (12), evab267. https://doi.org/10.1093/gbe/evab267
- Nicolini, F., Ghiselli, F., Luchetti, A. & Milani, L. (2023a) Bivalves as emerging model systems to study the mechanisms and evolution of sex determination: a genomic point of view. Genome Biology and Evolution, 15 (10), evad181. https://doi.org/10.1093/gbe/evad181
- Nicolini, F., Martelossi, J., Forni, G., Savojardo, C., Mantovani, B. & Luchetti, A. (2023b) Comparative genomics of Hox and ParaHox genes among major lineages of Branchiopoda with emphasis on tadpole shrimps. Frontiers in Ecology and Evolution, 11, 1046960. https://doi.org/10.3389/fevo.2023.1046960
- Novojilov, N.I. (1960) Podklass Gnathostraca. In: Chernysheva, N.E. (Ed.), Osnovy paleontologii. Chlenistonogie: trilobitoobraznye, i rakoobraznye. Gosudarstvennoe naučno-tehničeskoe izdatel’stvo literatury po geologii i ohrane nedr, Moskva, pp. 216–253.
- Oakley, T.H., Wolfe, J.M., Lindgren, A.R. & Zaharoff, A.K. (2013) Phylotranscriptomics to bring the understudied into the fold: Monophyletic Ostracoda, fossil placement, and pancrustacean phylogeny. Molecular Biology and Evolution, 30 (1), 215–233. https://doi.org/10.1093/molbev/mss216
- Olesen, J. (2000) An updated phylogeny of the Conchostraca: Cladocera clade (Branchiopoda, Diplostraca). Crustaceana, 73 (7), 869–886. https://doi.org/10.1163/156854000504877
- Olesen, J. (2009) Phylogeny of Branchiopoda (Crustacea)—character evolution and contribution of uniquely preserved fossils. Arthropod Systematics & Phylogeny, 67, 3–39. https://doi.org/10.3897/asp.67.e31686
- Olesen, J. & Richter, S. (2013) Onychocaudata (Branchiopoda: Diplostraca), a new high-level taxon in branchiopod systematics. Journal of Crustacean Biology, 33 (1), 62–65. https://doi.org/10.1163/1937240X-00002121
- Poschmann, M.J., Hegna, T.A., Astrop, T.I. & Hoffmann, R. (2024) Revision of Lower Devonian clam shrimp (Branchiopoda, Diplostraca) from the Rhenish Massif (Eifel, SW-Germany), and the early colonization of non-marine palaeoenvironments. Palaeobiodiversity and Palaeoenvironments, 104 (3), 535–569. https://doi.org/10.1007/s12549-023-00597-9
- Price, B.W., Winter, M., Brooks, S.J., Natural History Museum Genome Acquisition Lab, Darwin Tree of Life Barcoding collective, Wellcome Sanger Institute Tree of Life programme, Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective, Tree of Life Core Informatics collective & Darwin Tree of Life Consortium. (2022) The genome sequence of the blue-tailed damselfly, Ischnura elegans (Vander Linden, 1820). Wellcome Open Research, 7, 66. https://doi.org/10.12688/wellcomeopenres.17691.1
- Puttick M.N. (2019) MCMCtreeR: functions to prepare MCMCtree analyses and visualize posterior ages on trees. Bioinformatics, 35 (24), 5321–5322. https://doi.org/10.1093/bioinformatics/btz554
- Rambaut, A., Drummond, A.J., Xie, D., Baele, G. & Suchard, M.A. (2018) Posterior summarization in Bayesian phylogenetics using tracer 1.7. Systematic Biology, 67 (5), 901–904. https://doi.org/10.1093/sysbio/syy032
- Recknagel, H., Kamenos, N.A. & Elmer, K.R. (2018) Common lizards break Dollo’s law of irreversibility: genome-wide phylogenomics support a single origin of viviparity and re-evolution of oviparity. Molecular Phylogenetics and Evolution, 127, 579–588. https://doi.org/10.1016/j.ympev.2018.05.029
- Reis, M.D. & Yang, Z. (2011) Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Molecular Biology and Evolution, 28 (7), 2161–2172. https://doi.org/10.1093/molbev/msr045
- Richter, S., Olesen, J. & Wheeler, W.C. (2007) Phylogeny of Branchiopoda (Crustacea) based on a combined analysis of morphological data and six molecular loci. Cladistics, 23 (4), 301–336. https://doi.org/10.1111/j.1096-0031.2007.00148.x
- Savojardo, C., Luchetti, A., Martelli, P.L., Casadio, R. & Mantovani, B. (2019) Draft genomes and genomic divergence of two Lepidurus tadpole shrimp species (Crustacea, Branchiopoda, Notostraca). Molecular Ecology Resources, 19 (1), 235–244. https://doi.org/10.1111/1755-0998.12952
- Schwentner, M., Clavier, S., Fritsch, M., Olesen, J., Padhye, S., Timms, B.V. & Richter, S. (2013) Cyclestheria hislopi (Crustacea: Branchiopoda): a group of morphologically cryptic species with origins in the Cretaceous. Molecular Phylogenetics and Evolution, 66 (3), 800–810. https://doi.org/10.1016/j.ympev.2012.11.005
- Schwentner, M., Richter, S., Rogers, D.C. & Giribet, G. (2018) Tetraconatan phylogeny with special focus on Malacostraca and Branchiopoda: highlighting the strength of taxon-specific matrices in phylogenomics. Proceedings of the Royal Society B, 285 (1885), 20181524. https://doi.org/10.1098/rspb.2018.1524
- Simão, F.A., Waterhouse, R.M., Ioannidis, P., Kriventseva, E.V. & Zdobnov, E.M. (2015) BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31 (19), 3210–3212. https://doi.org/10.1093/bioinformatics/btv351
- Sun X.Y. & Cheng, J.H. (2023) Conflicts in mitochondrial phylogenomics of Branchiopoda, with the first complete mitogenome of Laevicaudata (Crustacea: Branchiopoda). Current Issues in Molecular Biology, 45 (2), 820–837. https://doi.org/10.3390/cimb45020054
- Sun, X.Y., Xia, X.H. & Yang, Q. (2016) Dating the origin of the major lineages of Branchiopoda. Palaeoworld, 25 (2), 303–317. https://doi.org/10.1016/j.palwor.2015.02.003
- Tasch, P. (1969) Branchiopoda. In: Moore, R.C. (Ed.), Treatise on Invertebrate Paleontology (Part R, Arthropoda 4 (1)). The Geological Society of America and The University of Kansas, Boulder, Colorado and Lawrence, Kansas, R128–R191.
- Uozumi, T., Ishiwata, K., Grygier, M.J., Sanoamuang, L.O. & Su, Z.H. (2021) Three nuclear protein-coding genes corroborate a recent phylogenomic model of the Branchiopoda (Crustacea) and provide estimates of the divergence times of the major branchiopodan taxa. Genes & Genetic Systems, 96 (1), 13–24. https://doi.org/10.1266/ggs.20-00046
- Van Damme, K. & Kotov, A.A. (2016) The fossil record of the Cladocera (Crustacea: Branchiopoda): Evidence and hypotheses. Earth-Science Reviews, 163, 162–189. https://doi.org/10.1016/j.earscirev.2016.10.009
- Van Damme, K., Cornetti, L., Fields, P.D. & Ebert, D. (2022) Whole-genome phylogenetic reconstruction as a powerful tool to reveal homoplasy and ancient rapid radiation in waterflea evolution. Systematic Biology, 71 (4), 777–787. https://doi.org/10.1093/sysbio/syab094
- Wersebe, M.J., Sherman, R.E., Jeyasingh, P.D. & Weider, L.J. (2023) The roles of recombination and selection in shaping genomic divergence in an incipient ecological species complex. Molecular Ecology, 32 (6), 1478–1496. https://doi.org/10.1111/mec.16383
- Wolfe, J.M., Daley, A.C., Legg, D.A. & Edgecombe, G.D. (2016) Fossil calibrations for the arthropod Tree of Life. Earth-Science Reviews, 160, 43–110. https://doi.org/10.1016/j.earscirev.2016.06.008
- Xu, S.L., Han, B.P., Martínez, A., Schwentner, M., Fontaneto, D., Dumont, H.J. & Kotov, A.A. (2021) Mitogenomics of Cladocera (Branchiopoda): Marked gene order rearrangements and independent predation roots. Molecular Phylogenetics and Evolution, 164, 107275. https://doi.org/10.1016/j.ympev.2021.107275
- Yang, Z. (2007) PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24 (8), 1586–1591. https://doi.org/10.1093/molbev/msm088
- Zhang, W.T., Chen, P.J. & Shen, Y.B. (1976) Fossil Conchostraca of China. Science Press, Beijing, 473 pp. [In Chinese]
- Zhang, F., Ding, Y.H., Zhou, Q.S., Wu, J., Luo, A.R. & Zhu, C.D. (2019) A high-quality draft genome assembly of Sinella curviseta: A soil model organism (Collembola). Genome Biology and Evolution, 11 (2), 521–530. https://doi.org/10.1093/gbe/evz013