Abstract
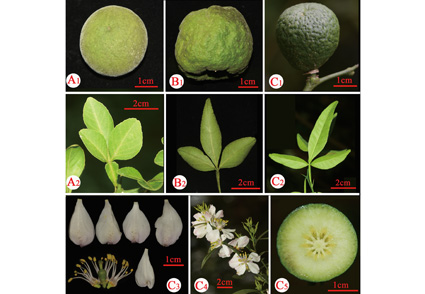
Citrus × pubinervia D. G. Zhang & Z. H. Xiang, Y. Wu, a new hybrid species is described and illustrated from central China. This new species is closely related to C. × polytrifolia in evergreen treelets and petiole with narrow wings but mainly differs from the latter by glabrous petals and glabrous fruits. The molecular phylogenetic analysis which based on four cpDNA (matK, rbcL-atpB, trnL-trnF, psbA-trnH) verifies the results of morphological analysis.
References
<p><em>Citrus</em> L. in GBIF Secretariat (2019) <em>GBIF Backbone Taxonomy.</em> Checklist dataset. https://doi.org/10.15468/39omei</p>
<p>Amar, M.H., Hassan, A.H.M., Biswas, M.K., Dulloo, E., Xie, Z.Z. & Guo, W.W. (2014) Maximum parsimony based resolution of interspecies phylogenetic relationships in <em>Citrus</em> L. (Rutaceae) using ITS of rDNA. <em>Biotechnology & Biotechnological Equipment</em> 28 (1): 61–67. https://doi.org/10.1080/13102818.2014.901665</p>
<p>Bayer, R.J., Mabberley, D.J., Morton, C., Miller, C.H., Sharma, I.K., Pfeil, B.E., Rich, S., Hitchcock, R. & Sykes, S. (2009) Molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. <em>American Journal of Botany</em> 96 (3): 668–685. https://doi.org/10.3732/ajb.0800341</p>
<p>Chen, Z.M. (1993) A new citrange from the alpine mountains of Western Hubei––Changyang citrange (<em>Poncirus trifoliata×Citrus ichangensis</em>?) [J]. <em>South China Fruits</em> 22 (04): 14. [in Chinese]</p>
<p>Ding, S.Q, Zhang, X.N, Bao, Z.R. & Ling, M.Q. (1984) A new species of <em>Poncirus</em> from China. <em>Acta Botanica Yunnanica</em> 6: 292–293.</p>
<p>Doyle, J.J. & Doyle, J.D. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. <em>Phytochemical Bulletin, Botanical Society of America</em> 19: 11–15.</p>
<p>Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. <em>Nucleic Acids Research</em> 32: 1792–1797. https://doi.org/10.1093/nar/gkh340</p>
<p>Fang, D.Q. (1993) Intra- and intergeneric relationships of <em>Poncirus polyandar</em>: Investigation by leaf isozymes. <em>Journal of Wuhan Botanical Research</em> 11 (1): 34–40.</p>
<p>Felsenstein, J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. <em>Evolution</em> 39: 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x</p>
<p>Gong, G.Z., Hong, Q.B., Peng, Z.C., Dong, J. & Xiang, S.Q. (2008) Genetic diversity of <em>Poncirus</em> and its phylogenetic relationships with relativesas revealed by nuclear and chloroplast SSR. <em>Acta Horticulturae Sinica</em> 35 (12): 1742–1750.</p>
<p>Linnaeus, C. (1753) Carl von (1753). <em>In: Species Plantarum</em> 2. 782 pp.</p>
<p>Linnaeus, C. (1763) <em>Species Plantarum</em>, Editio Secunda 2. 1101 pp.</p>
<p>Liu, L.Q., Yang, J. & Gu, Z.J. (2007) Karyomorphology and Taxonomic Position of <em>Poncirus polyandra</em> (Rutaceae). <em>Acta Botanica Yunnanica</em> 29 (2): 198–200.</p>
<p>Montrouzier, X. (1860) <em>Mémoires de l’Académie Nationale des Sciences, Belles-Lettres et Arts de Lyon: Classe des Sciences et Arts</em> 10: 186.</p>
<p>Morton, C.M. (2009) Phylogenetic relationships of the Aurantioideae (Rutaceae) based on the nuclear ribosomal DNA ITS region and three noncoding chloroplast DNA regions, atpb-rbcl spacer, rps16, and trnL-trnF. <em>Organisms Diversity & Evolution</em> 9 (1): 52–68. https://doi.org/10.1016/j.ode.2008.11.001</p>
<p>Muller, K., Muller, J. & Quandt, D. (2010) PhyDE-Phylogenetic Data Editor, version 0.9971. Available from: http://www.phyde.de (accessed 21 October 2021)</p>
<p>Nylander, J.A.A. (2004) <em>MrModeltest</em> v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.</p>
<p>Penjor, T., Anai, T., Nagano, Y., Matsumoto, R. & Yamamoto, M. (2010) Phylogenetic relationships of <em>Citrus</em> and its relatives based on rbcL gene sequences. Tree Genetics & Genomes 6 (6): 931–939. https://doi.org/10.1007/s11295-010-0302-1</p>
<p>Rafinesque, C.S. (1838) <em>Sylva Telluriana. Mantis synopt. New genera and species of trees and shrubs of North America, and other regions of the earth, omitted or mistaken by the botanical authors and compilers, or not properly classified, now reduced by their natural affinities to the proper natural orders and tribes.</em> 143 pp. https://doi.org/10.5962/bhl.title.248</p>
<p>Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L. Suchard, M.A. & Huelsenbeck, J.P. (2012) MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. <em>Systematic Biology</em> 61: 539–542. https://doi.org/10.1093/sysbio/sys029</p>
<p>Sang, T., Crawford, D.J. &, Stuessy, T.F. (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of <em>Paeonia </em>(Paeoniaceae)[J]. <em>American Journal of Botany</em> 84 (8):1120 –1121. https://doi.org/10.2307/2446155</p>
<p>Stamatakis, A. (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. <em>Bioinformatics</em> 22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446</p>
<p>Swingle, W.T. (1913) <em>Feroniella</em>, genre nouveau de la tribu des <em>citrea</em>, fonde nou le <em>F. oblata</em>, esplataou le de la ’Indo-Chine. <em>Bulletin de la Société Botanique de France</em> 59: 776. https://doi.org/10.1080/00378941.1912.10832512</p>
<p>Swingle, W.T. (1914) <em>Eremocitrus</em>, a new genus of hardy, drought-resistant citrous fruits from Australia. <em>Journal of Agricultural Research</em> 2: 85–100.</p>
<p>Swingle, W.T. (1915a) A new genus, <em>Fortunella</em>, comprising four species of kumquat oranges. <em>Journal of the Washington Academy of Sciences</em> 5: 165–176.</p>
<p>Swingle, W.T. (1915b) <em>Microcitrus</em>, a new genus of Australian citrous fruits. <em>Journal of the Washington Academy of Sciences</em> 5: 569–578.</p>
<p>Swingle, W.T. (1939) <em>Clymenia</em> and <em>Burkillanthus</em>, new genera; also three new species of <em>Pleiospermium </em>(Rutaceae-Aurantioideae). <em>Journal of the Arnold Arboretum</em> 20 : 250–263.</p>
<p>Turland, N.J., Wiersema, J.H., Barrie, F.R., Greuter, W., Hawksworth, D.L., Herendeen, P.S., Knapp, S., Kusber, W.-H., Li, D.-Z., Marhold, K., May, T.W., McNeill, J., Monro, A.M., Prado, J., Price, M.J. & Smith, G.F. (eds.) (2018) <em>International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017.</em> Regnum Vegetabile 159. Glashütten: Koeltz Botanical Books. https://doi.org/10.12705/Code.2018</p>
<p>Wu, G.A., Terol, J., Ibanez, V., López-García, A., Pérez-Román, E., Borredá, C., Domingo, C., Tadeo, F.R., Carbonell-Caballero, J., Alonso, R., Curk, F., Du, D.L., Ollitrault, P., Roose, M.L., Dopazo, J., Gmitter, F.G., Rokhsar, D.S. & Talon, M. (2018) Genomics of the origin and evolution of <em>Citrus</em>[J]. <em>Nature</em> 554 (7692): 1–20. https://doi.org/10.1038/nature25447</p>
<p>Zhang, D.X. & Mabberley, D.J. (2008) <em>Citrus</em>.<em> In: </em>Wu, Z. & Raven, P.H. (eds.) <em>Flora of China</em>, vol. 11. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis, pp. 90–96.</p>
<p>Zhang, Y., Gong, X. & Feng, X.Y. (2021) Phylogenetic position of <em>Poncirus</em> and <em>Poncirus polyandra</em> by DNA sequencing[J]. <em>Guihaia</em> 41 (01): 114–122.</p>
<p>Amar, M.H., Hassan, A.H.M., Biswas, M.K., Dulloo, E., Xie, Z.Z. & Guo, W.W. (2014) Maximum parsimony based resolution of interspecies phylogenetic relationships in <em>Citrus</em> L. (Rutaceae) using ITS of rDNA. <em>Biotechnology & Biotechnological Equipment</em> 28 (1): 61–67. https://doi.org/10.1080/13102818.2014.901665</p>
<p>Bayer, R.J., Mabberley, D.J., Morton, C., Miller, C.H., Sharma, I.K., Pfeil, B.E., Rich, S., Hitchcock, R. & Sykes, S. (2009) Molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. <em>American Journal of Botany</em> 96 (3): 668–685. https://doi.org/10.3732/ajb.0800341</p>
<p>Chen, Z.M. (1993) A new citrange from the alpine mountains of Western Hubei––Changyang citrange (<em>Poncirus trifoliata×Citrus ichangensis</em>?) [J]. <em>South China Fruits</em> 22 (04): 14. [in Chinese]</p>
<p>Ding, S.Q, Zhang, X.N, Bao, Z.R. & Ling, M.Q. (1984) A new species of <em>Poncirus</em> from China. <em>Acta Botanica Yunnanica</em> 6: 292–293.</p>
<p>Doyle, J.J. & Doyle, J.D. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. <em>Phytochemical Bulletin, Botanical Society of America</em> 19: 11–15.</p>
<p>Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. <em>Nucleic Acids Research</em> 32: 1792–1797. https://doi.org/10.1093/nar/gkh340</p>
<p>Fang, D.Q. (1993) Intra- and intergeneric relationships of <em>Poncirus polyandar</em>: Investigation by leaf isozymes. <em>Journal of Wuhan Botanical Research</em> 11 (1): 34–40.</p>
<p>Felsenstein, J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. <em>Evolution</em> 39: 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x</p>
<p>Gong, G.Z., Hong, Q.B., Peng, Z.C., Dong, J. & Xiang, S.Q. (2008) Genetic diversity of <em>Poncirus</em> and its phylogenetic relationships with relativesas revealed by nuclear and chloroplast SSR. <em>Acta Horticulturae Sinica</em> 35 (12): 1742–1750.</p>
<p>Linnaeus, C. (1753) Carl von (1753). <em>In: Species Plantarum</em> 2. 782 pp.</p>
<p>Linnaeus, C. (1763) <em>Species Plantarum</em>, Editio Secunda 2. 1101 pp.</p>
<p>Liu, L.Q., Yang, J. & Gu, Z.J. (2007) Karyomorphology and Taxonomic Position of <em>Poncirus polyandra</em> (Rutaceae). <em>Acta Botanica Yunnanica</em> 29 (2): 198–200.</p>
<p>Montrouzier, X. (1860) <em>Mémoires de l’Académie Nationale des Sciences, Belles-Lettres et Arts de Lyon: Classe des Sciences et Arts</em> 10: 186.</p>
<p>Morton, C.M. (2009) Phylogenetic relationships of the Aurantioideae (Rutaceae) based on the nuclear ribosomal DNA ITS region and three noncoding chloroplast DNA regions, atpb-rbcl spacer, rps16, and trnL-trnF. <em>Organisms Diversity & Evolution</em> 9 (1): 52–68. https://doi.org/10.1016/j.ode.2008.11.001</p>
<p>Muller, K., Muller, J. & Quandt, D. (2010) PhyDE-Phylogenetic Data Editor, version 0.9971. Available from: http://www.phyde.de (accessed 21 October 2021)</p>
<p>Nylander, J.A.A. (2004) <em>MrModeltest</em> v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.</p>
<p>Penjor, T., Anai, T., Nagano, Y., Matsumoto, R. & Yamamoto, M. (2010) Phylogenetic relationships of <em>Citrus</em> and its relatives based on rbcL gene sequences. Tree Genetics & Genomes 6 (6): 931–939. https://doi.org/10.1007/s11295-010-0302-1</p>
<p>Rafinesque, C.S. (1838) <em>Sylva Telluriana. Mantis synopt. New genera and species of trees and shrubs of North America, and other regions of the earth, omitted or mistaken by the botanical authors and compilers, or not properly classified, now reduced by their natural affinities to the proper natural orders and tribes.</em> 143 pp. https://doi.org/10.5962/bhl.title.248</p>
<p>Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L. Suchard, M.A. & Huelsenbeck, J.P. (2012) MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. <em>Systematic Biology</em> 61: 539–542. https://doi.org/10.1093/sysbio/sys029</p>
<p>Sang, T., Crawford, D.J. &, Stuessy, T.F. (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of <em>Paeonia </em>(Paeoniaceae)[J]. <em>American Journal of Botany</em> 84 (8):1120 –1121. https://doi.org/10.2307/2446155</p>
<p>Stamatakis, A. (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. <em>Bioinformatics</em> 22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446</p>
<p>Swingle, W.T. (1913) <em>Feroniella</em>, genre nouveau de la tribu des <em>citrea</em>, fonde nou le <em>F. oblata</em>, esplataou le de la ’Indo-Chine. <em>Bulletin de la Société Botanique de France</em> 59: 776. https://doi.org/10.1080/00378941.1912.10832512</p>
<p>Swingle, W.T. (1914) <em>Eremocitrus</em>, a new genus of hardy, drought-resistant citrous fruits from Australia. <em>Journal of Agricultural Research</em> 2: 85–100.</p>
<p>Swingle, W.T. (1915a) A new genus, <em>Fortunella</em>, comprising four species of kumquat oranges. <em>Journal of the Washington Academy of Sciences</em> 5: 165–176.</p>
<p>Swingle, W.T. (1915b) <em>Microcitrus</em>, a new genus of Australian citrous fruits. <em>Journal of the Washington Academy of Sciences</em> 5: 569–578.</p>
<p>Swingle, W.T. (1939) <em>Clymenia</em> and <em>Burkillanthus</em>, new genera; also three new species of <em>Pleiospermium </em>(Rutaceae-Aurantioideae). <em>Journal of the Arnold Arboretum</em> 20 : 250–263.</p>
<p>Turland, N.J., Wiersema, J.H., Barrie, F.R., Greuter, W., Hawksworth, D.L., Herendeen, P.S., Knapp, S., Kusber, W.-H., Li, D.-Z., Marhold, K., May, T.W., McNeill, J., Monro, A.M., Prado, J., Price, M.J. & Smith, G.F. (eds.) (2018) <em>International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017.</em> Regnum Vegetabile 159. Glashütten: Koeltz Botanical Books. https://doi.org/10.12705/Code.2018</p>
<p>Wu, G.A., Terol, J., Ibanez, V., López-García, A., Pérez-Román, E., Borredá, C., Domingo, C., Tadeo, F.R., Carbonell-Caballero, J., Alonso, R., Curk, F., Du, D.L., Ollitrault, P., Roose, M.L., Dopazo, J., Gmitter, F.G., Rokhsar, D.S. & Talon, M. (2018) Genomics of the origin and evolution of <em>Citrus</em>[J]. <em>Nature</em> 554 (7692): 1–20. https://doi.org/10.1038/nature25447</p>
<p>Zhang, D.X. & Mabberley, D.J. (2008) <em>Citrus</em>.<em> In: </em>Wu, Z. & Raven, P.H. (eds.) <em>Flora of China</em>, vol. 11. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis, pp. 90–96.</p>
<p>Zhang, Y., Gong, X. & Feng, X.Y. (2021) Phylogenetic position of <em>Poncirus</em> and <em>Poncirus polyandra</em> by DNA sequencing[J]. <em>Guihaia</em> 41 (01): 114–122.</p>